Chroma是Generate Bio开发一款蛋白设计算法,可以支持做骨架设计和序列设计。与Baker团队开发的RFdiffusion有一定相似之处。
开源代码: https://github.com/generatebio/chroma
论文地址:Illuminating protein space with a programmable generative model
基本功能测试
无条件采样(Unconditonal Sampling)
最基础的采样生成,随机生成结构,可以分为单体生成和复合物生成,适合用于随机结构数据集的构建。
1 | import appdirs |
指定二级结构采样(Secondary Structure Sampling)
算法支持指定每个氨基酸残基的二级结构(Helix为H,Sheet为E,loop为T)
1 | import appdirs |

符号形状采样(Symbol-like Structure Sampling)
这是一种炫技的蛋白结构设计,例如让设计蛋白结构像一个“G”。
但实测效果并没有那么像,论文中应该是选了典型case。
1 | import appdirs |
CATH采样(CATH-based Structure Sampling)
CATH编号是对蛋白二级结构一种编码,算法能够根据给定CATH编号来生成对应的二级结构。
这实际上上是在前面的二级结构采样基础上,增加了对CATH的查询解析。
1 | import appdirs |
提示词采样(Prompt-based Structure Sampling)
内嵌了LLM模型,用来支持基于给定提示词进行结构生成,
但实测效果不咋样,而且这样做的商业意义很有限。
1 | import appdirs |

全结构重采样(Full structure resampling)
算法可以做给定结构的重新采样,但采样效果与原始结构不像。
1 | import appdirs |
局部重采样(Partial structure resampling)
局部重新采样是指可以选择给定区域进行重新设计而保留其他区域。chroma通过一个类似pymol的selection语法进行结构的局部选择。重新采样设计的差异性较大。测试发现partial resampling其实是对局部结构的完全sampling,不能基于已有的局部结构,当然这可以作为固定receptor进行完全重新设计binder。
同时,测试使用了conditioners.ComposedConditioner([ss_conditioner, sub_conditioner])整合多个conditioner,
但发现会存在互相影响,
例如这里使用了生成指定二级结构类型的conditoner和指定结构区域的conditioner,
发现二级结构类型不好指定生成,生成结构和上面的类似。
1 | import appdirs |
Denovo Binder Design
Fc Fragment Binder design
1 | import appdirs |
通过构造pesudo-chain,利用conditioners.SubstructureConditioner可以实现针对给定receptor的binder生成,同时组合使用conditioners.ProClassConditioner可以引导生成特定类型的二级结构。如示例尝试对Fc生成一个40aa的binder,同时约束其为特定的二级结构,并使用colabfold进行验证。结果表明,chroma的骨架生成模型能生成较好的预期骨架,序列设计也能比较fit骨架,但是对docking pose的设计准确性欠佳(当然可能是colabfold不准)。
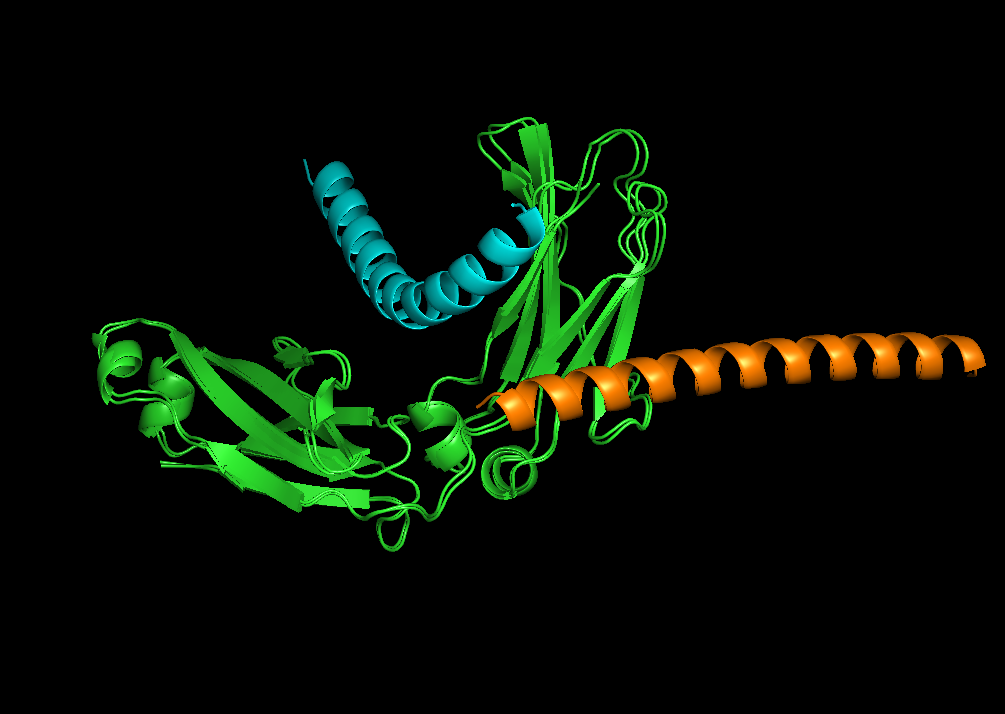
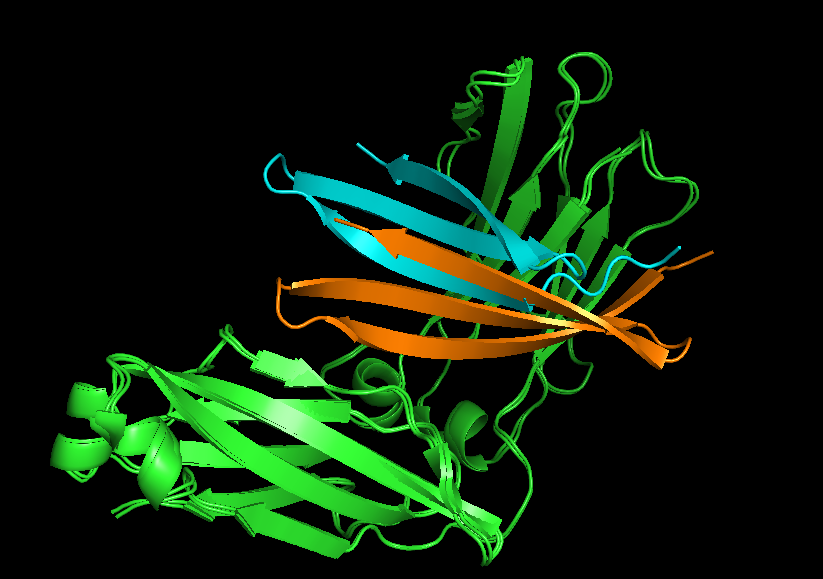
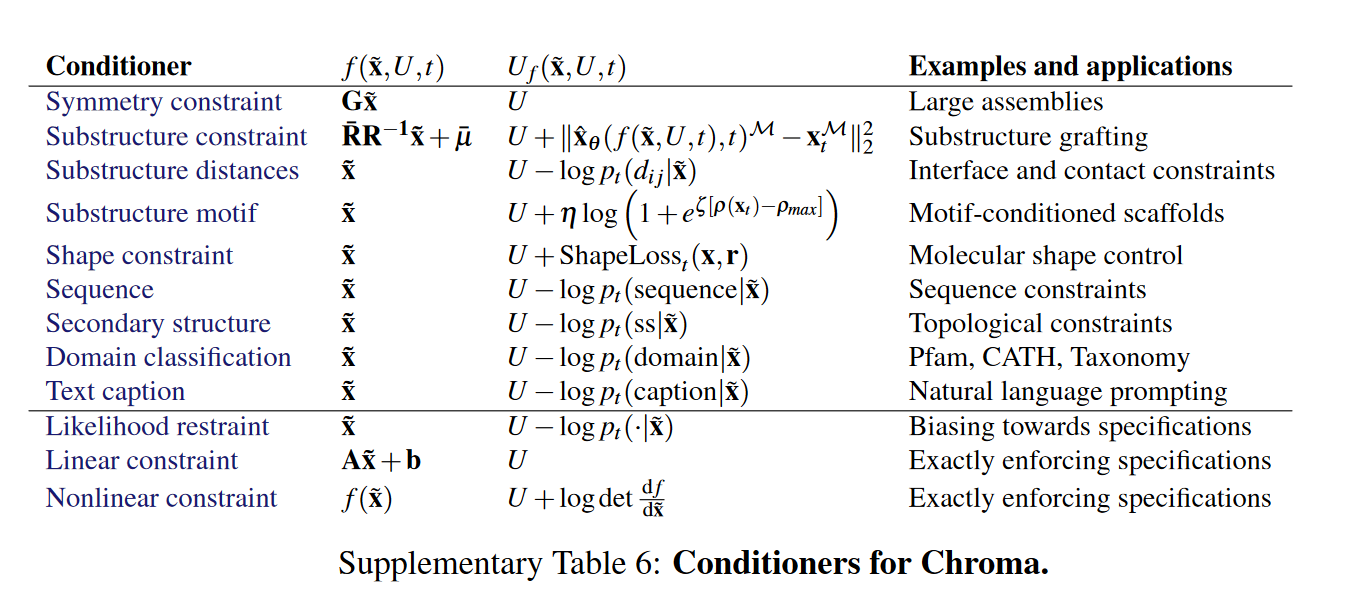
但目前的过程并没有任何对表位或者hotspot残基进行指定的设计,根据原文支持材料,应该是有一个substructure distances的conditioner可以完成这个实现,但在开源版本中并没有相关提供,不过根据这个conditioner开发指南和支持材料中给的逻辑推导,自行实现应该是具有可行度。
总结
Chroma是在RFdiffusion之后一个不错结构diffusion设计算法,但是在给定靶点和表位的设计上,还需要我们做更多技术性验证。